Introduction
Codi-Microscope is a next-generation nanoimaging and experimental analysis platform that empowers biologists and researchers to design, run, and visualize advanced experiments with unprecedented ease.
By integrating real-time imaging, experiment orchestration, and interactive visualization tools, Codi-Microscope bridges the gap between hardware-based microscopy and modern cloud-first research software. Researchers can focus on scientific discovery without being slowed down by complex imaging setups, scattered data pipelines, or computational barriers.
Key Features
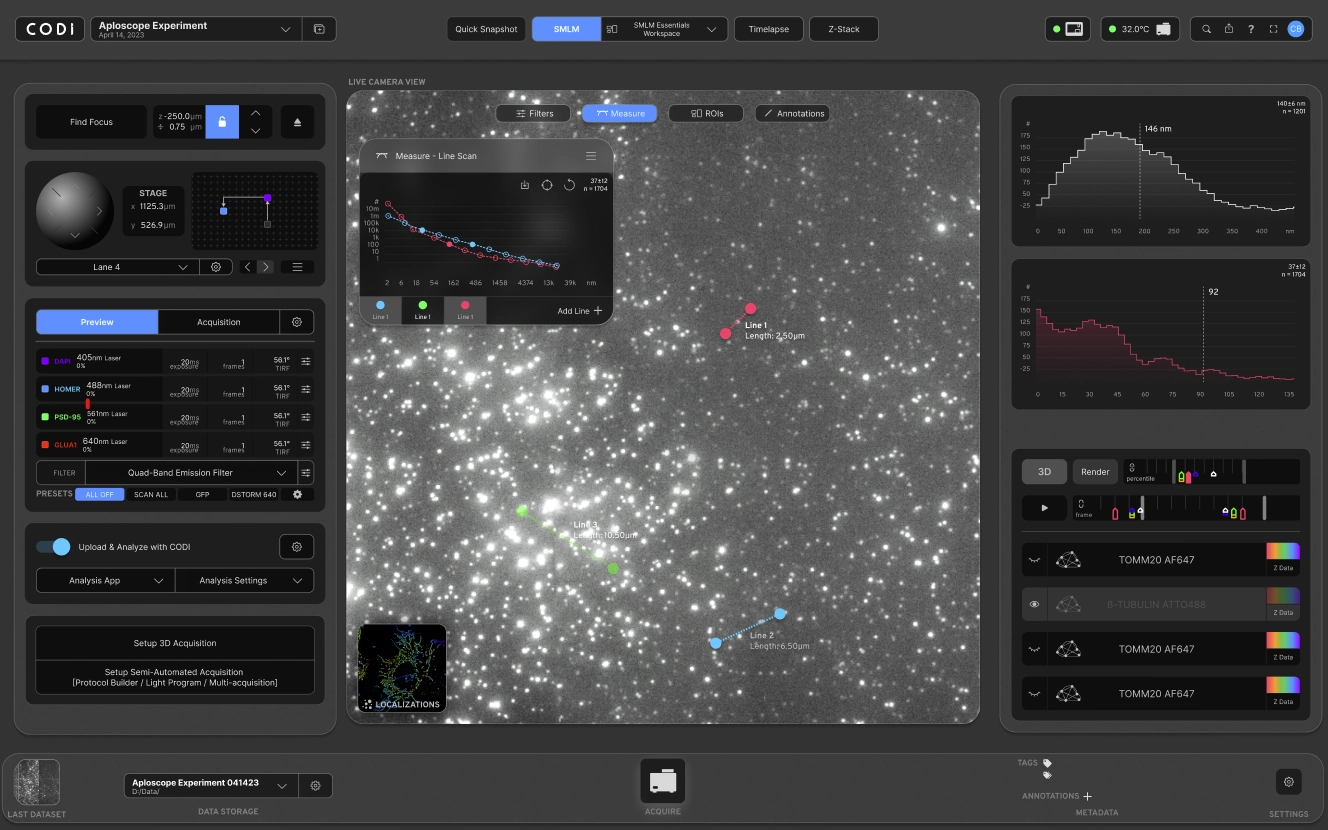
- Nanoimaging Control & Visualization: A browser-based interface for controlling microscopes, adjusting imaging parameters, and rendering nanoscale structures in real time using WebGL/Three.js.
- Experiment Orchestration: Define and automate experimental workflows, including multi-stage imaging, custom parameter tuning, and batch processing for high-throughput experiments.
- Image Analysis & Processing: Built-in Python-based algorithms for segmentation, alignment, and feature detection enable researchers to extract biological insights from raw images instantly.
- Data Management & Collaboration: Store large imaging datasets securely in the cloud, annotate results, and share projects with collaborators while maintaining reproducibility.
- Interactive Dashboards: Rich analytics and visualization dashboards give scientists insights into experiment performance, imaging quality, and biological measurements.
Technical Insights
- Frontend: Built with React, Next.js, and TypeScript, ensuring a highly responsive UI. TailwindCSS and Material UI provide a modern design system, while WebGL/Three.js powers interactive 2D and 3D nanoimaging visualizations.
- Backend & APIs: A Node.js backend manages experiment workflows and imaging requests, integrated with Python services that run image processing and machine learning pipelines. APIs are versioned and documented for seamless extensibility.
- Image Processing Engine: Python modules implement advanced bioinformatics and image analysis algorithms for segmentation, clustering, and feature detection, optimized for nanoscale biological data.
- Database & Persistence: Imaging datasets, metadata, and experiment logs are stored in PostgreSQL for relational reliability. Redis serves as a caching layer for frequently accessed imaging results and experiment states.
- Scalability & Deployment: Hosted on AWS, leveraging EC2 for compute, S3 for image storage, and RDS for managed databases. Dockerized microservices ensure reproducible deployments and easy scaling for multi-lab or institutional usage.
Challenges and Solutions
- Real-Time Nanoimaging Performance: Rendering high-resolution nanoimaging data in real time was computationally demanding. The solution involved GPU-accelerated rendering with WebGL and optimized data streaming pipelines for low-latency visualization.
- Large Data Volumes: Imaging experiments produce massive datasets, leading to storage and retrieval challenges. We addressed this by integrating AWS S3 with tiered storage policies and Redis caching for fast retrieval of recent results.
- Complexity of Experimental Workflows: Biologists often run multi-stage experiments with intricate parameters. To simplify this, we built a workflow orchestration engine that allows drag-and-drop experiment design, reducing manual setup and errors.
- Accessibility & Collaboration: Traditional microscopy tools are often locked to local machines. Codi-Microscope enables cloud-first access, allowing teams across labs and institutions to collaborate on shared experiments through secure project workspaces.